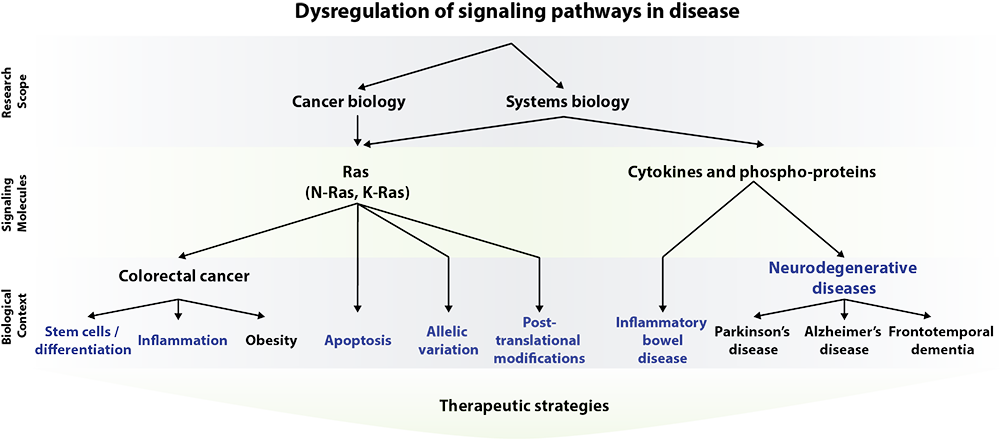
Our research can be broadly divided into two, although not mutually exclusive, areas: cancer biology and in vivo systems biology. The tree below shows the hierarchical structure of our research under the broad rubric of signaling pathway dysregulation.
Cancer Biology
Our cancer biology work focuses largely on the influence of activating Ras mutations in the pathogenesis of colorectal cancer. Our primary approach is to genetically manipulate Ras signaling pathways both in vivo (in the mouse colonic epithelium) and in vitro (in human colorectal cancer cells). We have developed mouse models of colon cancer that rely on mutationally activated K-Ras or N-Ras. Tumors from these mice mimic human colon cancers at both the molecular and histologic levels.
Using our model, we have found that mutant K-Ras and N-Ras contribute to tumorigenesis in different ways: while K-Ras locks colon cancer cells in a stem-like state and prevents them from differentiating, N-Ras suppresses stress-induced apoptosis. In further characterizing these models, our immediate goals are 1) to understand how activated K-Ras and N-Ras contribute to malignant progression in the colon; 2) to decipher the complex network of downstream signals that mediate their oncogenic phenotypes; and 3) to establish whether mutation of K-Ras or N-Ras confers sensitivity to specific targeted therapeutics. We hope these studies will translate into the identification of novel therapies to attack cancers expressing mutant Ras.
We are also exploring a novel mode regulation for Ras proteins that may constitute a new strategy for cancer therapy. We have found that Ras proteins are post-translationally acetylated on lysine 104. Acetylated Ras is locked into an inactive conformation, preventing it from promoting cancer. We are currently working to identify the enzymes that regulate the acetylation state of Ras. Drugs that alter the activity of these enzymes may be useful for cancer therapy.
in vivo Systems Biology
Our lab applies systems biology approaches to mouse models in an effort to understand genetically complex diseases. This work focuses on the development of computational models of dysregulated signaling networks in the context of disease. Currently, we are focusing on chronic inflammatory conditions of the gastrointestinal tract (inflammatory bowel diseases).
The novel aspect of our systems approach is that we focus strongly on the analysis of protein signaling data sets generated from in vivo experiments. These data sets are comprised of quantitative measurements of cytokine and phospho-protein levels.
Our ultimate goal is to elicit biologically/clinically meaningful information that will allow us to identify new therapeutic targets.